Drug Discovery - General Route
I recently stumbled upon a video made by the 'National Institutes of Allergy and Infectious Diseases' (NIAID) titled "How A Drug Becomes A Drug" which I will show below in a moment. Before I emphasize the importance of viewing the short video (less than 4 minutes), I want to introduce the agency NIAID -- which is a sub-agency of the 'National Institutes of Health'. Here is an excerpt describing the organization taken from the "Wikipedia" page for the "NIAID" below:
The National Institute of Allergy and Infectious Diseases (NIAID) is one of the 27 institutes and centers that make up the National Institutes of Health (NIH), an agency of the United States Department of Health and Human Services (HHS). NIAID's mission is to conduct basic and applied research to better understand, treat, and prevent infectious, immunologic, and allergic diseases.[1]NIAID has "intramural" (in-house) laboratories in Maryland and Montana, and funds research conducted by scientists at institutions in the United States and throughout the world. NIAID also works closely with partners in academia, industry, government, and non-governmental organizations in multifaceted and multidisciplinary efforts to address emerging health challenges such as the pandemic H1N1/09 virus.
The three main mission areas can be summarized from the "Wikipedia" page as follows:
Human Immunodeficiency Virus/Acquired Immunodeficiency Syndrome (HIV/AIDS)
The goals in this area are finding a cure for HIV-infected individuals; developing preventive strategies, including vaccines and treatment as prevention; developing therapeutic strategies for preventing and treating co-infections such as TB and hepatitis C in HIV-infected individuals; and addressing the long-term consequences of HIV treatment.
Biodefense and Emerging Infectious Diseases (BioD)
The goals of this mission area are to better understand how these deliberately emerging (i.e., intentionally caused) and naturally emerging infectious agents cause disease and how the immune system responds to them.
Infectious and Immunologic Diseases (IID)
The goal of this mission area is to understand how aberrant responses of the immune system play a critical role in the development of immune-related disorders such as asthma, allergies, autoimmune diseases, and transplant rejection. This research helps improve the understanding of how the immune system functions when it is healthy or unhealthy and provides the basis for development of new diagnostic tools and interventions for immune-related diseases.
The above mission covers every disease known and unknown. The National Institutes of Health is a huge organization made up of sub-agencies like the NIAID to divide up the mission. As such, the NIAID oversees the funding of drug research to a large extent in order to understand how infectious disease compromise the immune system -- the body at large. Additionally, the NIAID is interested in how drug discovery overcomes infectious diseases that have invaded our body. This includes the research behind the disease at the academic level.
I mentioned above a short video to highlight the general process of drug discovery from the academic level up all the way through to the consumer level -- i.e. the pharmacy. Here is the video below -- which is worth watching:
In the video above, the research is said to start at the basic science level at the university. This is true to an extent. Basic research into disease function and origin typically starts at the university level. Although, I would add that a fair amount of research is done at the industry level too by large drug companies. That research is typically targeted at a specific disease in which the pathway of progression or origin is known. I will explain more about the last sentence shortly.
The drug companies take the research done at the academic (university) level and carry the "small molecule" or "drug target" out to an actual therapeutic that is sold on the shelf of the pharmacy.
Why is this important to know?
Periodically, in the popular news, stories emerge about the over pricing of medication by companies like Turing pharmaceuticals (outrageous pricing) which cause wonder as to why such high prices exist for a given medication. These instances (of over pricing) are minimal compared to the price point needed to make a profit and move onto research more efficient drugs. The point is that research at the companies take time and money along with infrastructure.
The overall benefit of such research could be realized through an "open-access" network of drug targets and therapeutics (proprietary information at the moment) to which other researchers could access at their leisure. Arguments for such a system is that the funding has been provided by a government agency. Whereas arguments against such a system is loss of proprietary information. Tough call. Sorry for the divergence.
The goal of research is to find effective therapeutics (drugs) that treat a large part of the population. Side effects come about as a result of non-target delivery. The drug misses the target of intent or hits additional targets and causes extra problems. This is where the concept of "personalized medicine" comes in and will be discussed in future blog posts. For now, lets focus on designing drugs for a certain disease.
Drug Design 101
In order to design a drug to treat a certain disease or ailment, the pathology of the disease needs to be known. The origin of the disease needs to be known. How did the disease originate in the body?
Is the disease the result of a mutation in the genetic make-up of the person?
Is there a mutation in the DNA of the patient which causes a downstream mutation in the production of proteins?
Is the protein distorted in shape, contour which affects function?
Is the disease caused by an external agent (i.e. virus or bacteria)?
These problems can plague researchers success greatly for years. Luckily, over time, drug companies have built up libraries of "molecules" that serve as "messengers" or "therapeutics" that can hit a specific target that is involved in the process of the disease. Here is an excerpt from the "Wikipedia" page for "drug design" which I think will help you understand the process at the research level in either the university or industry setting:
Drug design, often referred to as rational drug design or simply rational design, is the inventive process of finding new medications based on the knowledge of a biological target.[1] The drug is most commonly an organic small molecule that activates or inhibits the function of a biomolecule such as a protein, which in turn results in a therapeutic benefit to the patient. In the most basic sense, drug design involves the design of molecules that are complementary in shape and charge to the biomolecular target with which they interact and therefore will bind to it. Drug design frequently but not necessarily relies on computer modeling techniques.[2] This type of modeling is sometimes referred to as computer-aided drug design. Finally, drug design that relies on the knowledge of the three-dimensional structure of the biomolecular target is known as structure-based drug design.[2] In addition to small molecules, biopharmaceuticals and especially therapeutic antibodies are an increasingly important class of drugs and computational methods for improving the affinity, selectivity, and stability of these protein-based therapeutics have also been developed.[3]The phrase "drug design" is to some extent a misnomer. A more accurate term is ligand design (i.e., design of a molecule that will bind tightly to its target).[4] Although design techniques for prediction of binding affinity are reasonably successful, there are many other properties, such as bioavailability, metabolic half-life, side effects, etc., that first must be optimized before a ligand can become a safe and efficacious drug. These other characteristics are often difficult to predict with rational design techniques. Nevertheless, due to high attrition rates, especially during clinical phases of drug development, more attention is being focused early in the drug design process on selecting candidate drugs whose physicochemical properties are predicted to result in fewer complications during development and hence more likely to lead to an approved, marketed drug.[5] Furthermore, in vitro experiments complemented with computation methods are increasingly used in early drug discovery to select compounds with more favorable ADME (absorption, distribution, metabolism, and excretion) and toxicological profiles.[6]
The drug designer is looking for a "biological target" that is involved in either the origin or progression of the disease. As mentioned above, there are two popular processes: computer-aided drug based design and structure-based drug design. Both involve information on the biological target of interest.
What might a biological target look like?
A biological target can vary in definition depending on the nature of the disease. For instance, if the disease involves the distortion or mutation of a protein, then the surface of the protein would be considered the biological target. Specifically, the site of interest for a drug to interact with is referred to as the "active site." Here is an image take from the "Wikipedia" page for "Active Site" to help assist the reader in what that might look like:
Source: Thomas Shafee - Own work, CC
As you can see, the protein appears to be like a "blob" in the image above. The reason for that to emphasize the "binding site" or "catalytic site" not the overall structure which is of little concern to the drug designer. Remember that proteins are made up of amino acids which in turn form large "macromolecules" some of which are referred to as Proteins. I wrote an earlier blog about oligosaacharides are made up of simple sugars.
Starting from the picture above, now, the video below might make sense to watch before we proceed with our discussion of drug design. The video is titled "A Basic Introduction to Drugs, Drug Targets, and Molecular Interactions" and is just over 4 minutes long -- and definitely worth watching.
The video above is more technical than some readers might want to view in order to understand the process. Therefore, we should back off a little on the "technical side" and focus on the "development" side of drug development -- from a simplistic standpoint.
Are you ready to understand drug design from a simple standpoint?
Alright, here we go!
In order to do so, I decided to borrow a few slides from a recent webinar offered online by the American Chemistry Society. The webinar was titled "Crystallography As A Drug Design And Delivery Tool" and was given by Dr. Vincent Stoll of AbbVie -- where he serves as the Director of Structural Biology.
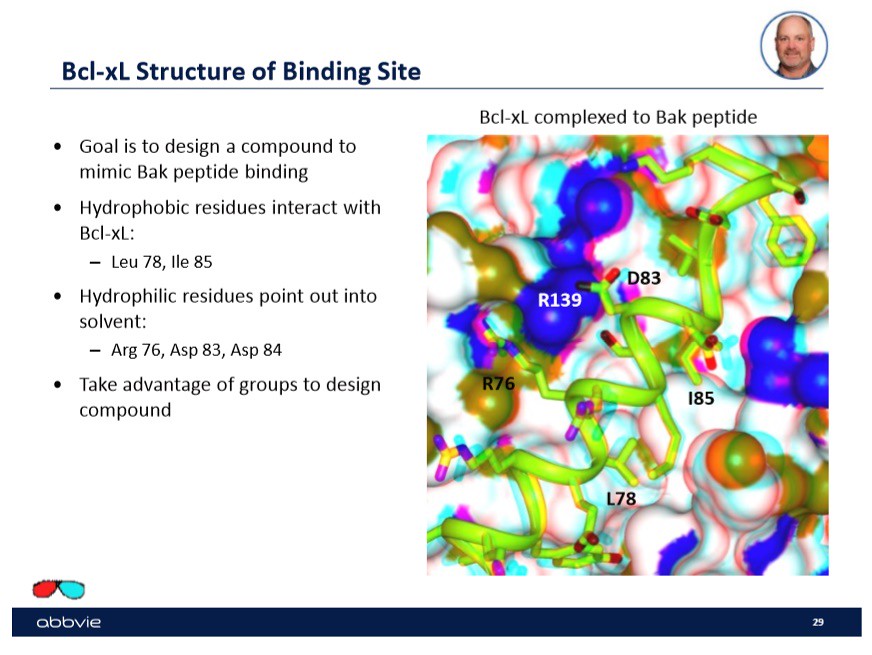
One of the examples that Dr. Stoll used to talk about drug design was binding to the transmembrane molecule B-Cell-Lymphoma-Extra-Large or bcl-xl in the mitochondria. In his talk, he focused on a few binding sites shown in the slide below with a picture:
Specifically, in this case the company wanted to design a drug candidate that would "mimic" the peptide Bak binding. Shown to the right on the slide are the sites or "active sites" that the peptide Bak bind to on the transmembrane molecule bcl-xl. In order to find a drug that will mimic the binding of the peptide, the drug will have to have the ability to bind to multiple sites on the transmembrane molecule.
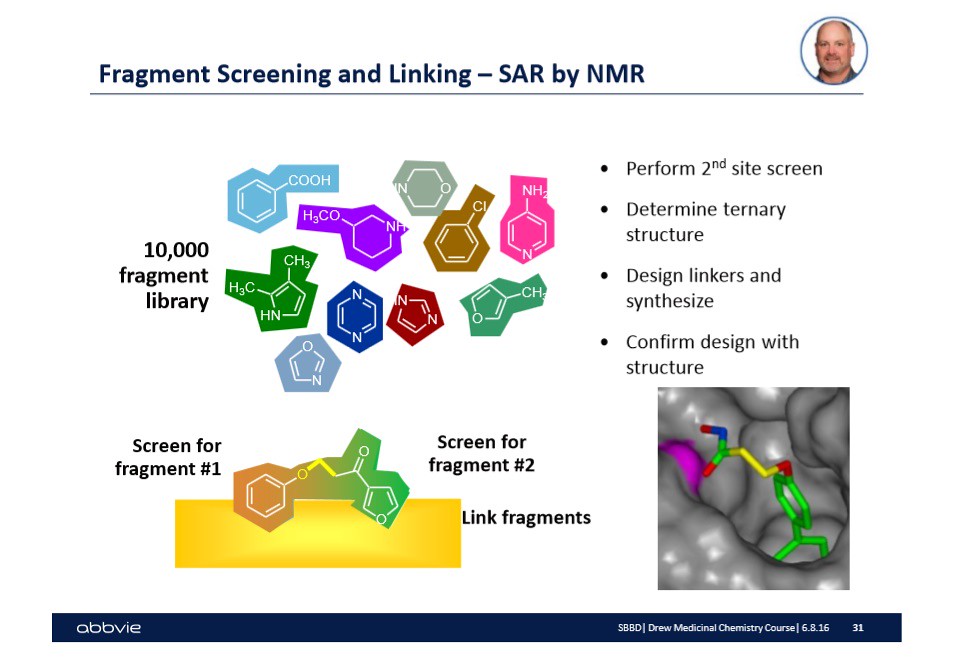
Fortunately, over time, large drug companies have built up a data base or library of 'molecules' that will bind to similar or exact sites. In the slide below, I show a yellow surface with two molecules hovering above the surface -- slightly bound -- taken from Dr. Stoll's talk:
There is a lot of information on the slide shown above. Let me walk you through the relevant information for drug design 101. First, I mentioned that each drug company kept a library or database with a bunch of 'fragments' that are intended to hit specific targets on biological surfaces. These biological surfaces can be viewed as the picture shown to the right in the slide. They might be a protein surface, or another biological surface of interest to drug manufacturers.
In the case above, the two molecules shown on the yellow surface -- one is a brown color while the other is a green color. The different color is to illustrate that the molecules are fragments designed to hit a specific type of target or active site on a biological surface. In this case, the biological surface is the transmembrane molecule bcl-xl.
Once the fragments have been identified that will occupy and hit the desired targets or active sites, then the challenge is to link the fragments together by chemistry. This step in of itself is often challenging and does not guarantee that the newly formed molecule (of two fragments with a linker molecule) will work. Therefore, in the picture above, there are possible linker molecules that exist within the pharmaceutical database that have been shown to work in other cases.
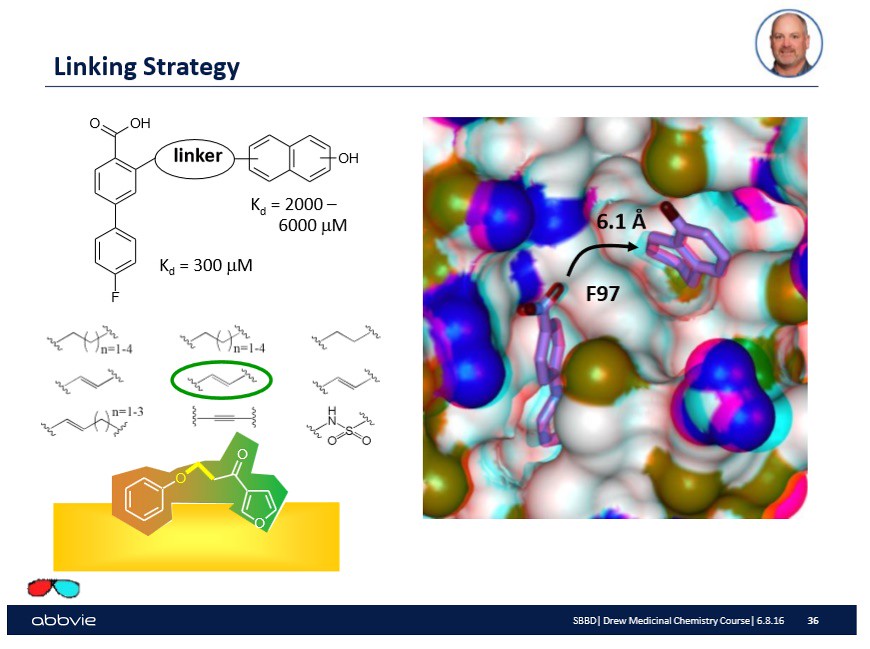
After linking the two fragments together, the next step is to verify by spectroscopy that the total or linked molecule worked.
How is this accomplished?
In the lab, the substrate or biological surface will have a drop of the linked molecule injected onto the surface. Then the surface which should have the drug bound to the active sites will be investigated using a spectroscopic technique like Nuclear Magnetic Resonance Spectroscopy. Upon confirmation, a number will be reported as shown on the slide that indicates the binding affinity of the molecule onto the surface:
In the slide above, there are a couple of numbers reported that make sense to drug designers but probably not the reader -- you. Do not worry. Over time (through other blog posts) you will come to understand their meaning. What is important to understand at this point is that after linking molecular fragments together, an experiment occurs to understand if the drug or linked molecule is as effective as the fragments are alone.
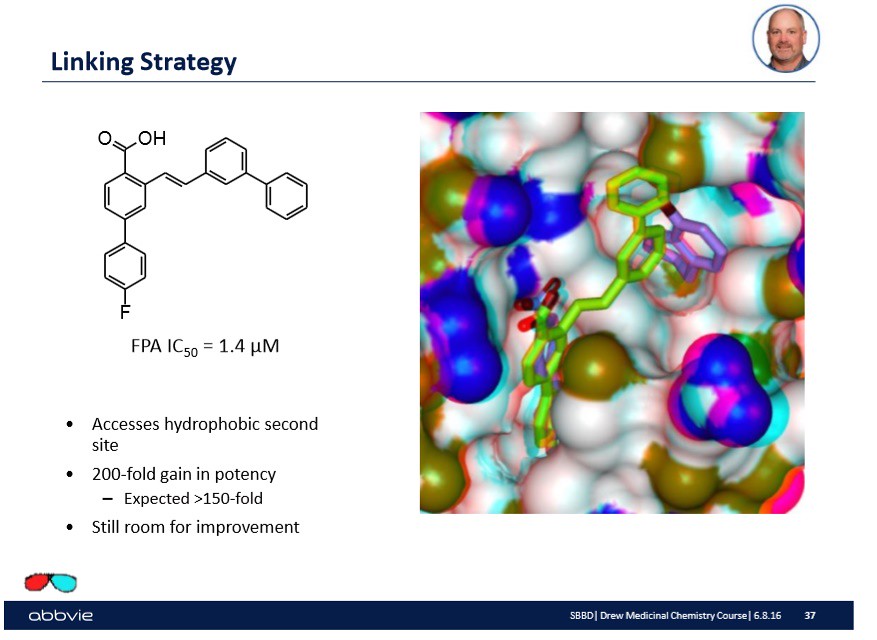
Furthermore, the pharmaceutical company might understand the chemistry of the active site to a large extent and further modify the linked molecules to make a more "potent" drug or linked molecule. On the slide below, I show from Dr. Stoll's talk such a modification:
Again, the overall take home message is that the molecular modification done to the linked molecule has some effect.
Is that effect better or worse?
Can there be a further modification to the linked molecule or now drug to enhance the ability to mimic the peptide binding?
Who knows. That is why research is continuously pushed forward and costs money to find out.
Conclusion...
In the above paragraphs, my intention was to introduce briefly the process of drug design. As we speak though, changes are being made to parts of the process. Outsourcing of linker molecules is occurring as are mergers and acquisitions of large companies by even larger pharmaceutical companies. Which potentially means that the shared database or libraries of available drug targets is growing. The process is dynamic but slow at the same time.
Discovering the mechanisms of disease and cures as a result is the dream of every drug designer. Progress is unfortunately slowed down by the trial and error process. Research takes time and money to complete. Furthermore, improvements to existing drugs take time. I will leave you with another short video about the progression of the medical research field:
Until next time, have a great day!!!!